Joint multi-ancestry and admixed GWAS reveals the complex genetics behind human cranial vault shape
- Apr 30, 2024
- 3 min read
By: Seppe Goovaerts, Hanne Hoskens, Ryan J. Eller, Noah Herrick, Anthony M. Musolf, Cristina M. Justice, Meng Yuan, Sahin Naqvi, Myoung Keun Lee, Dirk Vandermeulen, Heather L. Szabo-Rogers, Paul A. Romitti, Simeon A. Boyadjiev, Mary L. Marazita, John R. Shaffer, Mark D. Shriver, Joanna Wysocka, Susan Walsh, Seth M. Weinberg & Peter Claes

Goovaerts, S. et al. Joint multi-ancestry and admixed GWAS reveals the complex genetics behind human cranial vault shape. Nat Commun 14, 7436 (2023).
This work represents the first comprehensive analysis of the genetics underlying human cranial vault morphology. Although the vault – the portion of the cranium surrounding the neocortex – is an anatomical region of great interest to numerous fields of study (e.g., anthropology and evolutionary biology, neuroscience, forensics, and clinical genetics among others), our understanding of the genetic factors driving variation in its size and shape are poorly understood. Much like our prior work on human facial genetics (White et al. Nature Genetics, 2021; 53:45-53), this work is important because elucidating the factors behind the visible variation present among humans – the traits that endow each of us with our unique appearance – is fundamental to understanding who we are as individuals and the diversity we observe around us every day.
By employing an innovative phenotyping pipeline coupled with a multivariate Genome-wide association study (GWAS) in a multi-ancestry cohort comprising high levels of admixture, we identified 30 statistically significant loci and then independently replicated 20 of these signals. We show that genes at these signals are enriched for relevant developmental processes, are differentially expressed in mouse cranial bones, and play a role in vault dysmorphology. For example, using recently available WGS data, we show that several of our nominated SNPs are also associated with forms of non-syndromic craniosynostosis, thereby genetically linking normal-range and dysmorphic cranial vault variation. We accomplish this by integrating data from a multitude of sources (patient data, mouse tissues, cell lines, and embryonic tissues) analyzed through diverse bioinformatics approaches (RNAseq, ChIPseq, Gene Ontology analysis) to provide an extensive and in-depth documentation of the genetic architecture of the cranial vault.
Large-scale collection of 3D craniofacial images is a huge challenge, hence, we had to rely on pre-existing neuroimaging datasets. Extracting cranial vault shape from structural MRIs, necessitates extensive image processing. The computational resources offered by the VSC significantly cut down on the time needed to do this and ultimately made this study possible
Furthermore, we demonstrate innovative and novel approaches to handle some of the emerging challenges in contemporary genetics research. One such example is the inclusion of admixed individuals in GWAS, who are often omitted, even from recent multi-ancestry GWASs that meta-analyze results from single-ancestry cohorts. Another example relates to the capture of biologically relevant traits from large imaging datasets, particularly when those images were collected and optimized for another purpose (e.g., brain studies). Here, by means of an innovative image processing pipeline, we tackle complex phenotyping from MRIs, which contain vast amounts of information and are becoming increasingly available from large-scale biobanks.
Key findings:
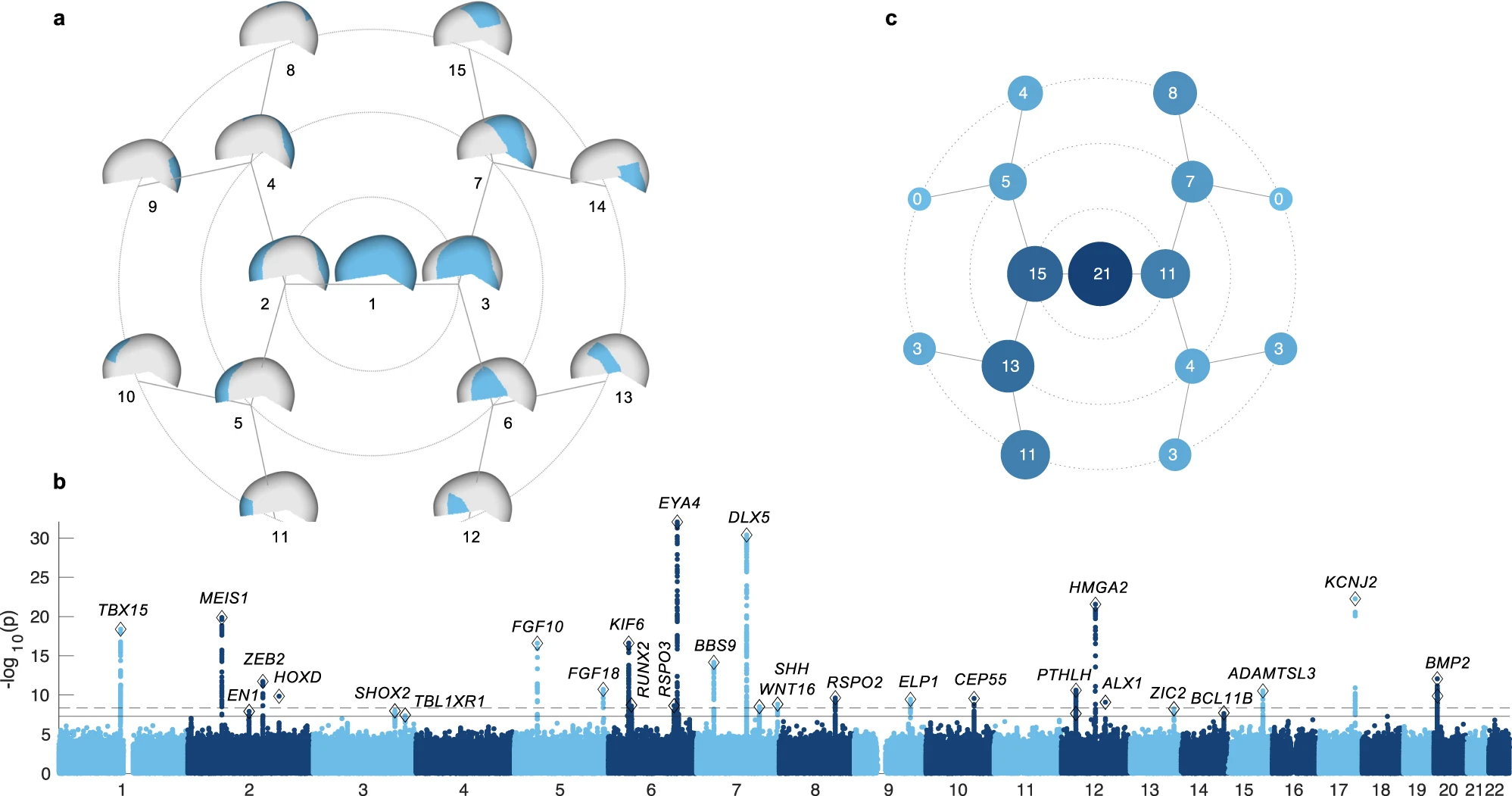
30 genomic loci were associated with cranial vault shape (Fig 1b), 29 of these were novel. Among the implicated genes were RUNX2, a master regulator of intramembranous ossification, the process through which the bones of the cranial vault form, along with several genes directly related to RUNX2.
Some genetic variants affect global cranial vault shape, while others have more local effects, e.g., in the frontal bone or forehead (Supplementary data 1).
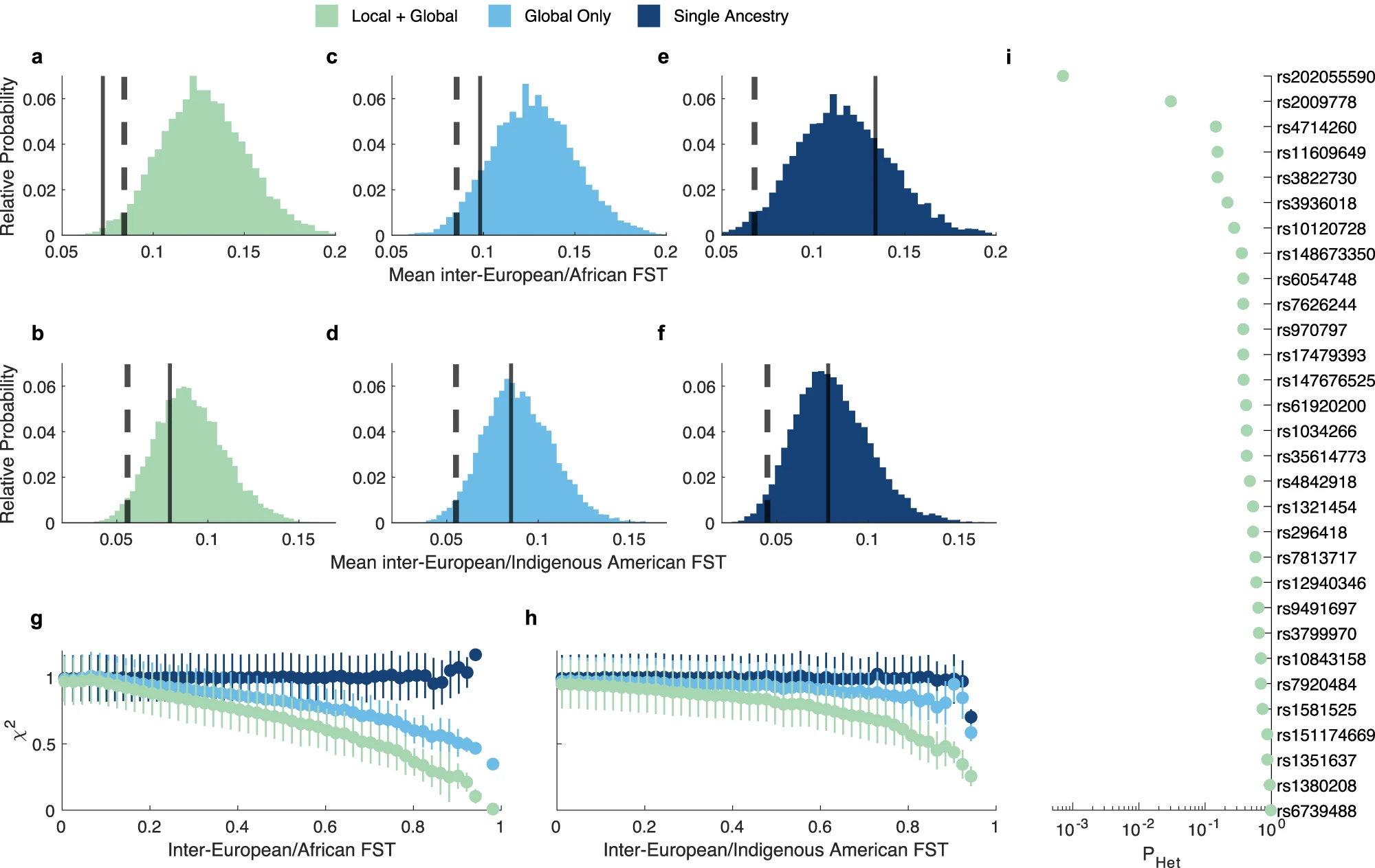
The identified genetic variants have mostly homogeneous effects across ancestries (Fig 2i and Supplementary data 2)
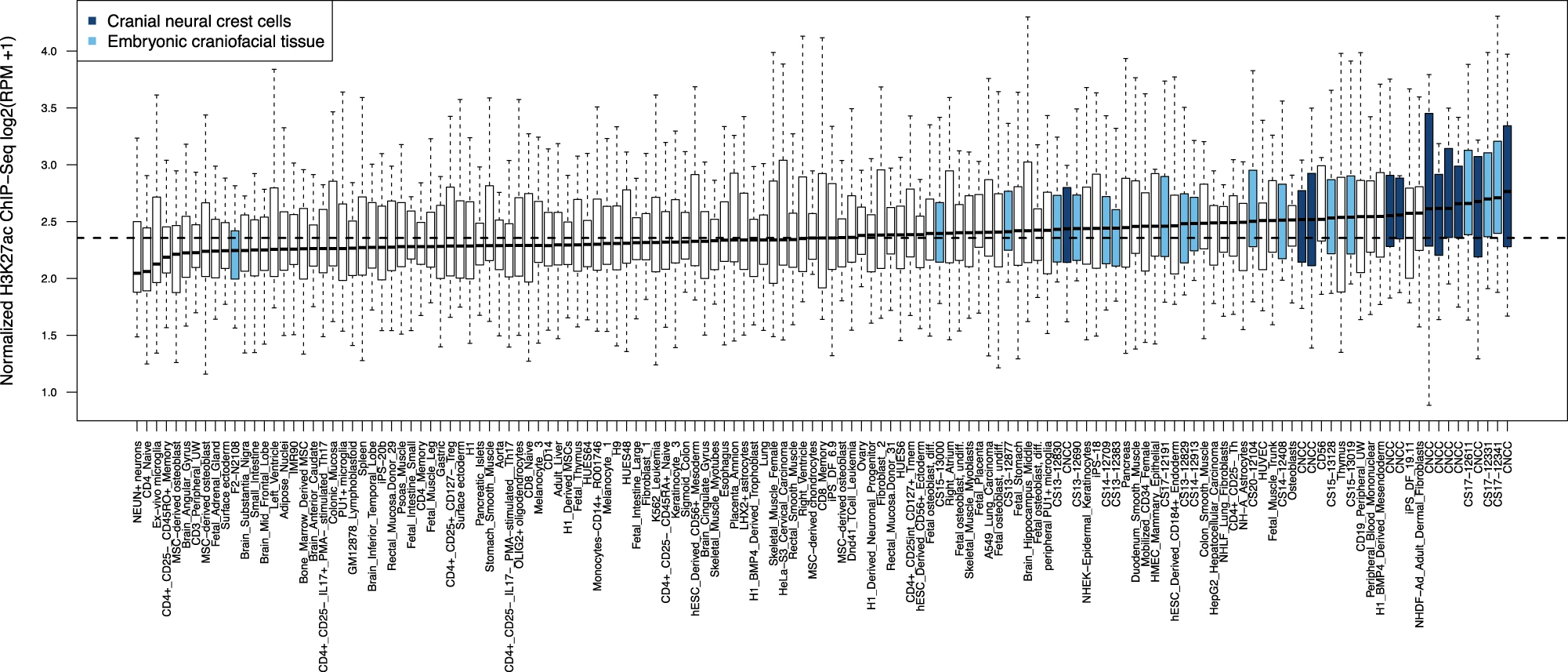
The identified genomic regions are preferentially active in cranial neural crest cells and embryonic craniofacial tissue, in other words, early during embryonic development (Fig 3).
Several genes implicated in our GWAS were differentially expressed across the frontal and parietal bone in mouse embryos. Genes including ALX1 were more active in the frontal bone, which also were they showed the strongest association in our GWAS.
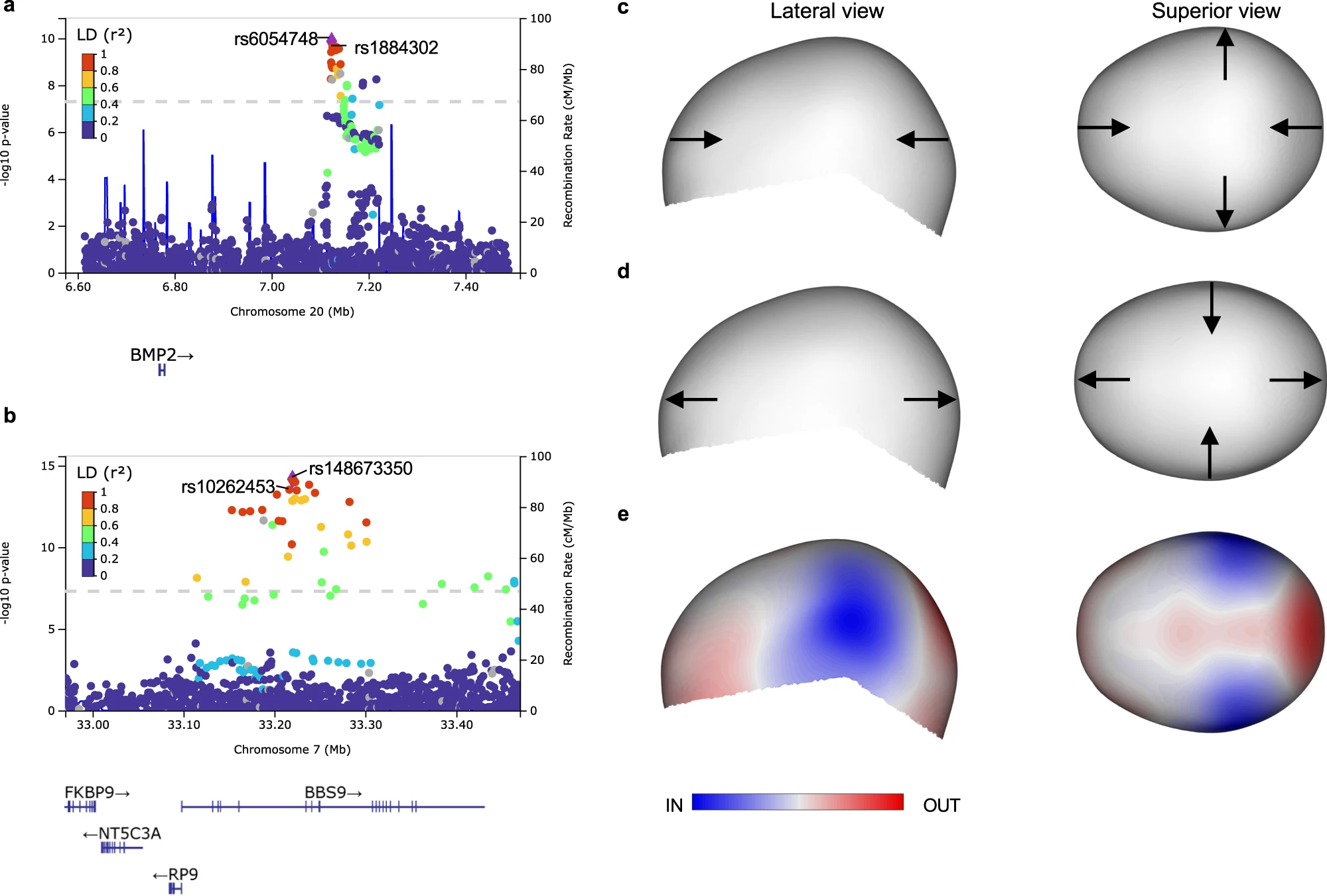
In small cohorts of single-suture craniosynostosis, three of the variants associated with normal-range cranial vault shape variation were also significantly associated with risk for craniosynostosis, i.e., near the genes BMP2, BBS9, and ZIC2. The effect of our variant near BMP2 (in the healthy cohort) agreed with the cranial vault deformation seen in sagittal craniosynostosis. (Fig 4)
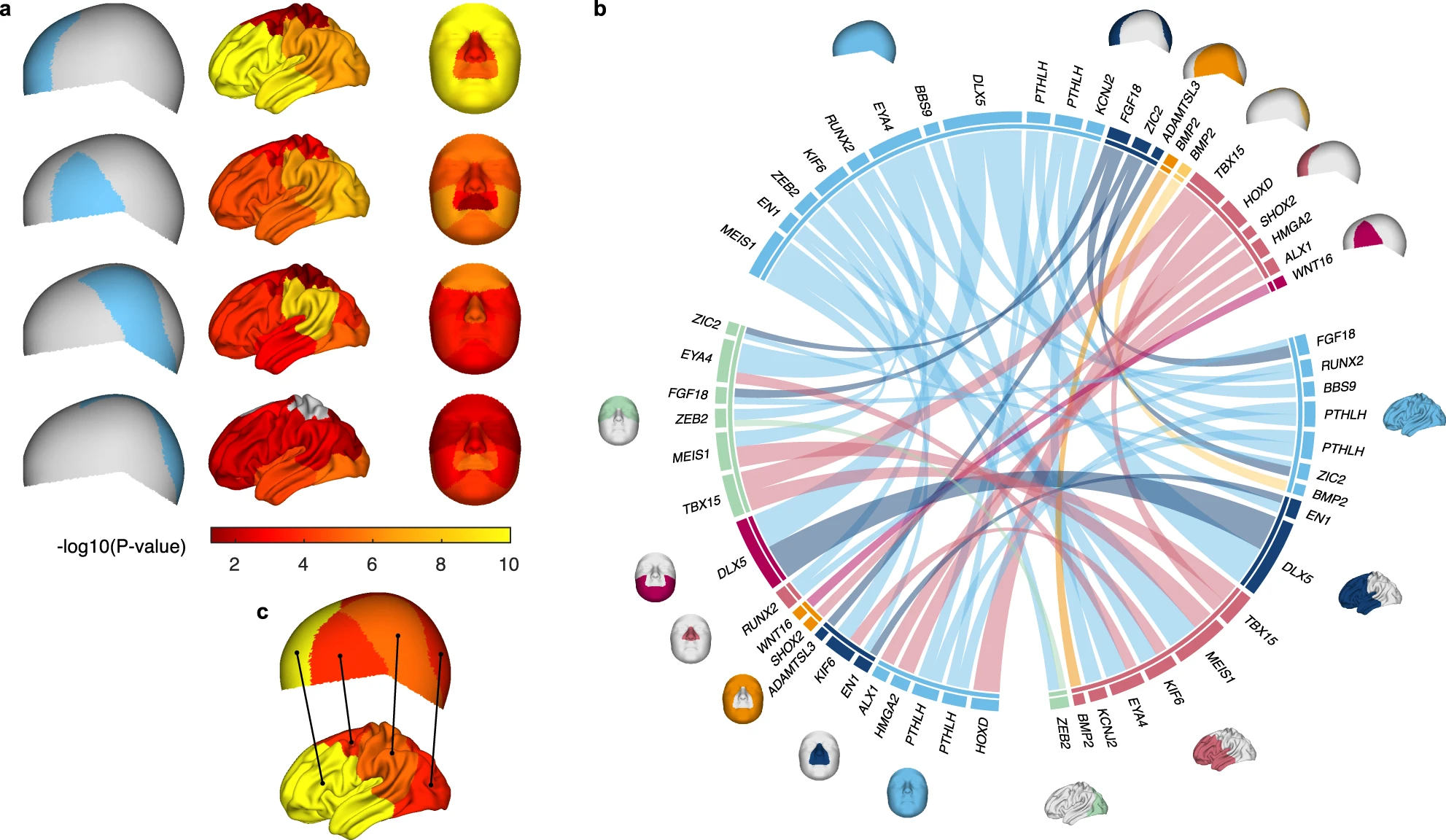
Many of variants identified for the cranial vault are also associated with shape variation of the face and brain. (Fig 5)
Read the full publication in the Nature Portfolio here